Welcome to GPCR-PCA
GPCR-PCA (G protein coupled receptor properties in pan-cancers), a web-based tool to deliver fast and customizable functionalities based on cancer sequencing analysis results. It offers notable features including expression, hazard ratio, G protein signaling pathway and mRNA structure of GPCRs in pan-cancers. Meanwhile, it offers GPCR drugs/ligands, G4 structures on promoter and super-enhancer/enhancer for GPCRs for user-defined query.
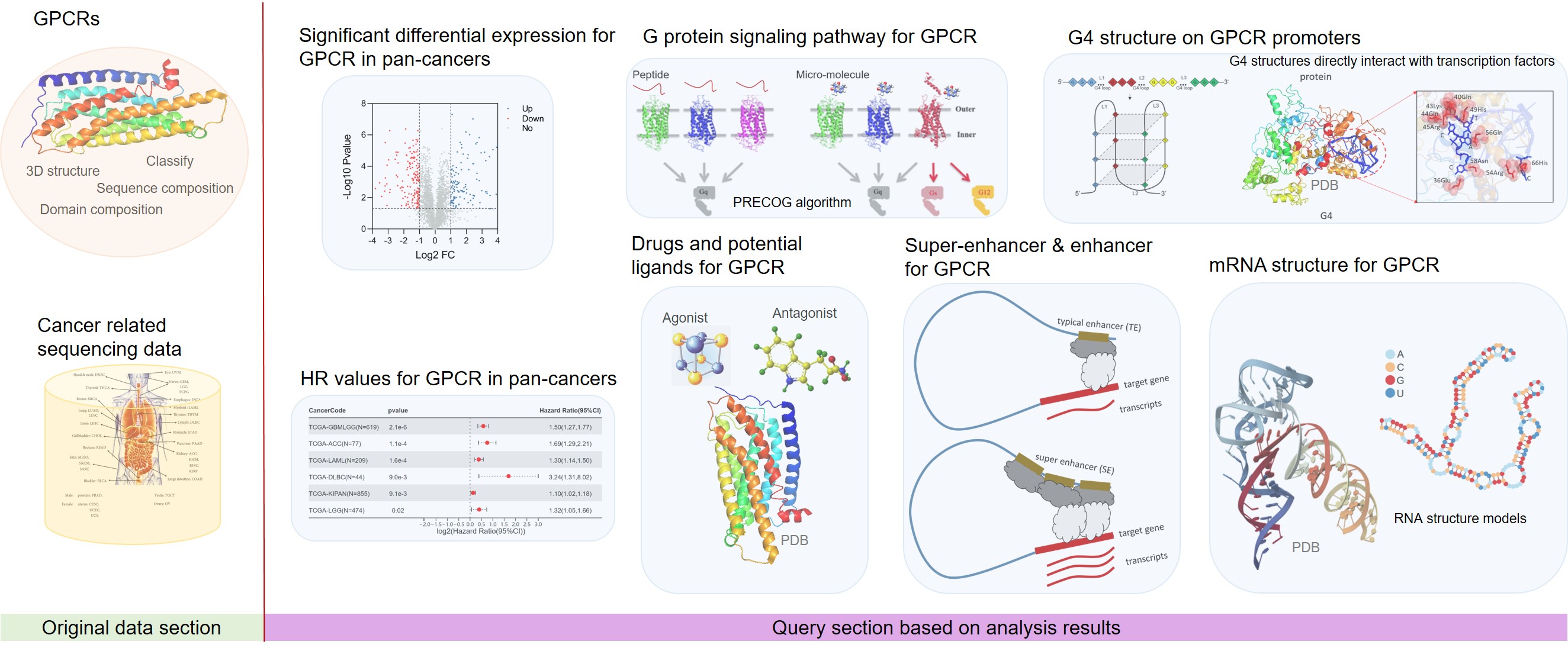
Based on big data and artificial intelligence algorithms, we are committed to summarizing the analysis results of these sequencing data to provide important research information and medication guidance for biomedical users.
GPCR information
GPR35
ADGRF3
See details for more genes.
Details
*The sort of amino acid composition: A, C, D, E, F, G, H, E, I, K, L, M, N, P, Q, R, S, T, V, W, Y, Others.
**The sequence of amino acid numbers in the domain: N-term, TMH1, ICL1, TMH2, ECL1, TMH3, ICL2, TMH4, ECL2, TMH5, ICL3, TMH6, ECL3, TMH7, C-term.
- represents unknown.
Significant differential expression for GPCR in cancers
GPR160
CXCR4
See details for more genes.
Details
ACC: Adrenocortical carcinoma,BLCA: Bladder Urothelial Carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL: Cholangiocarcinoma, COAD: Colon adenocarcinoma, COADREAD: Colon adenocarcinoma/Rectum adenocarcinoma Esophageal carcinoma, DLBC: Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, ESCA: Esophageal carcinoma, FPPP: FFPE Pilot Phase II FFPE, GBM: Glioblastoma multiforme,GBMLGG: Glioma, HNSC: Head and Neck squamous cell carcinoma, KICH: Kidney Chromophobe, KIPAN: Pan-kidney cohort (KICH+KIRC+KIRP), KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute Myeloid Leukemia, LGG: Brain Lower Grade Glioma, LIHC: Liver hepatocellular carcinoma, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma,MESO: Mesothelioma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PCPG: Pheochromocytoma and Paraganglioma, PRAD: Prostate adenocarcinoma, READ: Rectum adenocarcinoma, SARC: Sarcoma, SKCM: Skin Cutaneous Melanoma, STAD: Stomach adenocarcinoma, STES: Stomach and Esophageal carcinoma, TGCT: Testicular Germ Cell Tumors, THCA: Thyroid carcinoma, THYM: Thymoma, UCEC: Uterine Corpus Endometrial Carcinoma, UCS: Uterine Carcinosarcoma, UVM: Uveal Melanoma.
HR (hazard ratio) values for GPCR in pan-cancers
HTR2C
CXCR6
See details for more genes.
Details
ACC: Adrenocortical carcinoma,BLCA: Bladder Urothelial Carcinoma, BRCA: Breast invasive carcinoma, CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL: Cholangiocarcinoma, COAD: Colon adenocarcinoma, COADREAD: Colon adenocarcinoma/Rectum adenocarcinoma Esophageal carcinoma, DLBC: Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, ESCA: Esophageal carcinoma, FPPP: FFPE Pilot Phase II FFPE, GBM: Glioblastoma multiforme,GBMLGG: Glioma, HNSC: Head and Neck squamous cell carcinoma, KICH: Kidney Chromophobe, KIPAN: Pan-kidney cohort (KICH+KIRC+KIRP), KIRC: Kidney renal clear cell carcinoma, KIRP: Kidney renal papillary cell carcinoma, LAML: Acute Myeloid Leukemia, LGG: Brain Lower Grade Glioma, LIHC: Liver hepatocellular carcinoma, LUAD: Lung adenocarcinoma, LUSC: Lung squamous cell carcinoma,MESO: Mesothelioma, OV: Ovarian serous cystadenocarcinoma, PAAD: Pancreatic adenocarcinoma, PCPG: Pheochromocytoma and Paraganglioma, PRAD: Prostate adenocarcinoma, READ: Rectum adenocarcinoma, SARC: Sarcoma, SKCM: Skin Cutaneous Melanoma, STAD: Stomach adenocarcinoma, STES: Stomach and Esophageal carcinoma, TGCT: Testicular Germ Cell Tumors, THCA: Thyroid carcinoma, THYM: Thymoma, UCEC: Uterine Corpus Endometrial Carcinoma, UCS: Uterine Carcinosarcoma, UVM: Uveal Melanoma.
G protein signaling pathway (predicted) for GPCR
OR2M5
OR2A25
See details for more genes.
Details
G-proteins: Gs (GNAS and GNAL), Gi/o (GNAI1, GNAI2, GNAI3, GoA, GoB and GNAZ), Gq/11 (GNA11, GNA14, GNA15 and GNAQ), G12/13 (GNA12 and GNA13), and β-arrs (β-arr1-GRK2, β-arr2 and β-arr2-GRK2).
Drugs and potential ligands for GPCR
GPR35
PTH1R
See details for more genes.
Details: FDA approaved drugs for GPCR
Details: DrugBank ids for GPCR
Drug id details
Drug details were obtained from FDA (https://www.fda.gov/) and Drugbank(https://go.drugbank.com/).
G4 structure on promoters
ACKR1
ACKR2
See details for more genes.
Details
G4 represents G-quadruplex. 0 represents unknown.
*G4 in cancer cell/tissue were obatined based on G4 sequencing.
**PQS represents potential quadruplex sequence, which is obatined by motif exploring based on reference genome hg19.
293T: human embryonic kidney 293T cells; A549: human pulmonary carcinoma A549; BRCA: Breast Cancer tissue; HeLa: human cervical carcinoma cell; HepG2: hepatoma cell line; K562: Leukemia K562 cell.
Super-enhancer & typical enhancer for GPCR in cancers
ADORA2B
GPRC5A
See details for more genes.
Details for SE (super enhancer)
Details for TE (typical enhancer)
SE represents super enhancer. TE represents typical enhancer.
Super Enhancers and typical enhancers in cancer cells/tissues were obatined based on public H3K27ac chip-seq data. MACS V2.0 was used to calculated H3K27ac chip-seq peaks, which were regarded as typical enhancers. ROSE with default parameters was used to calculated the accurate postion of super enhancers.
A498:human kidney cancer cells; A549:human pulmonary carcinoma A549; BCPAP:Human thyroid cancer cells; Bladder tumor; breast cancer; GBM:glioblastoma; HCT116:Human colorectal adenocarcinoma cells; HEK293: human embryonic kidney 293 cells; HeLa:human cervical carcinoma cell; HN12: Squamous cell carcinoma of the head and neck; OV: Ovarian serous cystadenocarcinoma; PRAD: Prostate adenocarcinoma; pancreatic cancer.
mRNA structure for GPCR in cancers
ADORA2B
GPRC5A
See details for more genes.
Details
To profile the structurome of pre-mRNAs for GPCR genes, we used the ViennaRNA package (v2.4.18) to in silico predict potential short- and long-range duplexes formed between pairwise 150-nt windows along pre-mRNA for each GRCR gene.
GM12878: human lymphoblastoid cell line; H1: H1 Human embryonic stem cell line; HeLa: human cervical carcinoma cell; HepG2: hepatoma cell line, hNPC:Human neural precursor cells; IMR90:Human foetal lung cells; and K562: Leukemia K562 cell.
Search customized GPCR gene in all functionalities
About us
If you have any questions about the original data, operation or theory of the website, please refer to our paper or contact us by email.
Yue Hou:houyue@xjtu.edu.cn
Chuanjun Shu: chuanjunshu@njmu.edu.cn
Cite Us
…….
Acknowledgement
We would like to thank other members in our lab for discussion and suggestions.
This work was supported by the Natural Science Foundation of Jiangsu Province (No.BK20220313), Natural Science Research Project of Colleges and Universities in Jiangsu Province (No.22KJB180004), China Postdoctoral Science Foundation (2021M692581), and Natural Science Basic Research Program of Shaanxi (2021JQ-023).